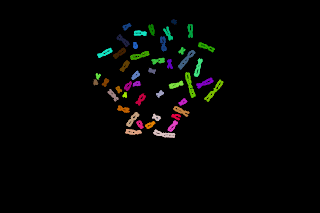Telomeres quantification can be done at low magnification in interphasic cells. A microscopic field was recorded with a CCD camera at low magnification (objective x25). Nuclei were counterstained with DAPI and the telomeres were detected with a Cy3-PNA probe.
From the field image, a nucleus is segmented and the DAPI and Cy3 spectral components are isolated. Even at low magnification, the telomeric spots can be segmented as follow:
Background is removed by top-hat filtering with a small structuring element (a disk of size=5). The top-hat image is then submitted to a hmin operator, leaving the peaks higher than 200 gray level units, then regional maxima are found.
The regional maxima are labelled to obtain markers for the watershed algorithm. Once thresholded, the top hat image is converted into distance map using euclidian or chess-board distance without significant difference.
Watershed doesn't succeed very well in separating spots belonging to the lower cluster of three spots. It may be interesting to use a high pass filtered image to build a distance map.
The segmented spots can be overlaid to the original image (blue:telomeric spots, yellow:lines separating the spots after watershed). The quantification can be then performed on the original image (left) but the background must be estimated. The quantification can also be done on the Top-Hat filtered image where the background value is close to zero.
To quantify the spots, two parameters are measured: the spot area and the mean of the spot. The spot intensity is set to the product of the area by the mean gray level value (without taking the exposure time into account).
The area, mean and intensity values can be mapped onto an image or displayed as a histogram (from left to right: histogram of spots mean values, of spots area and histogram of spots intensity).
The following python script relies on pymorph, mahotas and scipy. It could be tested with skimage.
The following python script relies on pymorph, mahotas and scipy. It could be tested with skimage.
# -*- coding: utf-8 -*-
"""
Created on Fri Dec 16 13:14:14 2011
@author: Jean-Patrick Pommier
"""
import scipy.ndimage as nd
import pylab as plb
import numpy as np
import mahotas as mh
import pymorph as pm
import skimage as sk
import skimage.morphology as mo
def E_distanceTransform(bIm):
#from pythonvision.org
dist = nd.distance_transform_edt(bIm)
dist = dist.max() - dist
dist -= dist.min()
dist = dist/float(dist.ptp()) * 255
dist = dist.astype(np.uint8)
return dist
def Chess_distanceTransform(bIm):
#from pythonvision.org
dist = nd.distance_transform_cdt(bIm,metric='chessboard')
dist = dist.max() - dist
dist -= dist.min()
dist = dist/float(dist.ptp()) * 255
dist = dist.astype(np.uint8)
return dist
def gray12_to8(im):
i=0.062271062*im
return pm.to_uint8(i)
path="/home/claire/Applications/ImagesTest/JPP50/16/"
CC="DAPI/"
PR="CY3/"
im="1.TIF"
dapi=mh.imread(path+CC+im)
cy3=mh.imread(path+PR+im)
#==============================================================================
# Extracting known nuclei
#==============================================================================
nuc=dapi[215:264,1151:1198]
telo=cy3[215:264,1151:1198]
#==============================================================================
# Telomeres Segmentation with pymorph and ndimage
#==============================================================================
op=pm.open(telo,pm.sedisk(5))
top=telo-op
h=pm.hmin(top,200)
remax=pm.regmax(h)
blur=nd.gaussian_filter(h,3)
hi=h-1.0*blur
mask=np.copy(hi)
hi[mask<0]=0
remax=pm.edgeoff(remax)
markers,n=nd.label(remax)
print n," spots detected"
distE=E_distanceTransform(top>200)
distC=Chess_distanceTransform(top>200)
segE,L=pm.cwatershed(distE,markers,return_lines=True)
segC,L=pm.cwatershed(distC,markers,return_lines=True)
#==============================================================================
# Display segmentation on one nucleus
#==============================================================================
print 'display'
RedNuc=mh.bwperim((nuc>1000),n=4)
BlueSpot=mh.bwperim((segE>0),n=4)
GreenSpot=mh.bwperim((segC>0),n=4)
displayTelo=pm.overlay(gray12_to8(telo),red=RedNuc,blue=BlueSpot,yellow=L)
displayTeloC=pm.overlay(gray12_to8(top),red=RedNuc,green=GreenSpot)
#==============================================================================
# Quabtification
#==============================================================================
mean=pm.grain(top,segE,'mean','data')
print np.shape(mean)
area=pm.blob(segE,'area','data')
meanIm=pm.grain(top,segE,'mean','image')
areaIm=pm.blob(segE,'area','image')
print mean
plb.figure(1)
plb.subplot(141,frameon=False, xticks=[], yticks=[])
plb.title('raw 12bits')
plb.imshow(telo)
plb.subplot(142,frameon=False, xticks=[], yticks=[])
plb.title('top Hat')
plb.imshow(top)
plb.subplot(143,frameon=False, xticks=[], yticks=[])
plb.title('hmin')
plb.imshow(h)
plb.subplot(144,frameon=False, xticks=[], yticks=[])
plb.title('regional max')
plb.imshow(remax)
plb.figure(2)
plb.subplot(331,frameon=False, xticks=[], yticks=[])
plb.title('markers')
plb.imshow(markers)
plb.subplot(332,frameon=False, xticks=[], yticks=[])
plb.title('Thresholded Top-Hat')
plb.imshow(top>200)
plb.subplot(334,frameon=False, xticks=[], yticks=[])
plb.title('Euclid dist map ')
plb.imshow(distE)
plb.subplot(335,frameon=False, xticks=[], yticks=[])
plb.title('watershed seg')
plb.imshow(segE)
plb.subplot(333,frameon=False, xticks=[], yticks=[])
plb.title('TopH+hi pass')
plb.imshow(hi)
plb.subplot(337,frameon=False, xticks=[], yticks=[])
plb.title('Chess d map')
plb.imshow(distC)
plb.subplot(338,frameon=False, xticks=[], yticks=[])
plb.title('watershed Chess')
plb.imshow(segC)
plb.figure(3)
plb.subplot(121,frameon=False, xticks=[], yticks=[])
plb.title('telo+seg')
plb.imshow(displayTelo)
plb.subplot(122,frameon=False, xticks=[], yticks=[])
plb.title('telo+seg')
plb.imshow(displayTeloC)
plb.figure(4)
plb.subplot(231,frameon=False, xticks=[], yticks=[])
plb.title('Quant image=mean')
plb.imshow(meanIm)
plb.subplot(232,frameon=False, xticks=[], yticks=[])
plb.title('Quant img=area')
plb.imshow(areaIm)
plb.subplot(233,frameon=False, xticks=[], yticks=[])
plb.title('Quant img=mean*area')
plb.imshow(areaIm*meanIm)
plb.subplot(234)
plb.hist(mean)
plb.subplot(235)
plb.hist(area)
plb.subplot(236)
Q=areaIm*meanIm
plb.hist(Q.flatten()[Q.flatten()>0])
plb.show()