Up to now, image labelling was performed with seeds chosen manually and drawn with the gimp or ImageJ.
The seeds localization have to be found automatically and mathematical morphology can provide solutions. The chromosomal axis is close to the particule squeleton:
From the binary particules, imagej gives the following distance map:
with the following code recorded from the macro:
and then the squeleton. From the squeleton, peculiar points are searched: branching points and end-points using a binary hit-and-miss operator:
End points detection with imagej:
Good candidates are selected.
The seeds localization have to be found automatically and mathematical morphology can provide solutions. The chromosomal axis is close to the particule squeleton:
 | |
| Binary chromosomes |
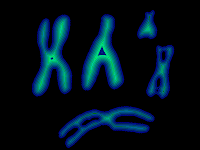 | |
| Distance map of binary particules (false colors) |
- run("Make Binary");
- run("Distance Map");
- run("3-3-2 RGB");
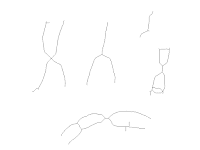 |
| Binary particules squeleton |
End points detection with imagej:
- run("Duplicate...", "title=squel.png");
- run("Make Binary");
- run("Skeletonize");
- run("BinaryHitOrMiss ", "kernel_a=[0 0 0 0 1 0 0 1 0 ] rotations=[rotate 45] white");
- run("Dilate");
- run("Select All");
- run("Copy");
- selectWindow("bin.png");
- run("Paste");
- run("Duplicate...", "title=EndPoints-Triple.png");
- run("Make Binary");
- run("Skeletonize");
- run("BinaryHitOrMiss ", "kernel_a=[0 1 0 1 1 1 2 2 2 ] rotations=[rotate 45] white");
- run("Dilate");
- selectWindow("bin.png");
- selectWindow("EndPoints-Triple.png");
- run("Select All");
- run("Copy");
- selectWindow("bin.png");
- run("Paste");
- saveAs("PNG", "/home/jeanpat/Applications/Images_test/png/essai07/ImageJ_HitnMiss_pointstriples_type2.png");
- open("/home/jeanpat/Applications/Images_test/png/essai07/ImageJ_HitnMiss_pointstriples.png");
- run("Select All");
- run("Copy");
- selectWindow("ImageJ_HitnMiss_pointstriples_type2.png");
- run("Paste");
 | |
| end-points (black), branching-points (colored) |
























