Chromosomes segmentation is the first step of MFISH or QFISH analysis.
Let's try to identify chromosomes from a DAPI stained metaphase:
The image was acquired with a 12 bits CCD camera. Microscopic field illumination is supposed homogeneous and no flat-field correction will be performed.
To segment the chromosomes, first check that Gabriel Landini's morpholological plugins is installed, and load the image with ImageJ. ImageJ indicates that the image is now a 16 bits image.
The interphasic nuclei yield saturated pixels at 4095 in the image histogram :
The gray level value of the first peak corresponding to the backgroung is around 300. Let's substract this value to the image and then perform a local background substraction with a rolling ball value set to 15 using the Hilo LUT to visualize the background.
Now the image histogram yields:
Erase the pieces of nuclei with the eraser tool. Threshold the image interactively and label the binary image then display it with the 3-3-3 RGB LUT:
This procedure yields under segmented chromosomes. From the original binary image. By performing a watershed and relabel the new image, we get:
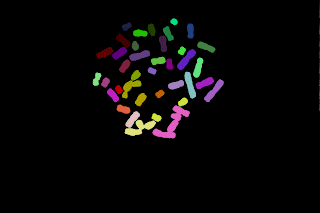
The touching chromosomes are now well separated but most chromosomes are now over segmented, anyway the result may be interesting since the parts obtained grossly corresponds to p and q chromosomal arms.
To get a good segmentation, an interactive step with the drawing tool may be here necessary:
Another example of chromosomes segmentation without manual modification (that is without painting tools) leading to over segmentation:
Here identifying first the chromosomes help to choose parameters to perform chromosomes segmentation. Some iterations between image segmentation and the chromosomes recognition step must be introduced to automatise the process.





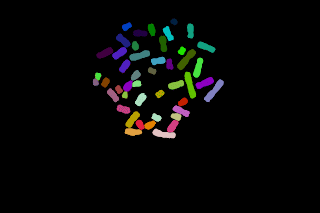
No comments:
Post a Comment