Introduction
Dicentric chromosomes are the signature of recombination events which may imply telomeric motifs. To observe dicentric chromosomes in senescent cells, a cell culture followed by an enrichment step is performed to get enough metaphasic cells. However metaphasic cells become difficult to get as the cell culture reaches senescence or if cells differenciate, thus most of fusions between short telomeres, when they occur, remain hidden in interphasic cells.
Telomeric fusions can generate head to head TTAGGG motifs as:
(TTAGGG)n(CCCTAA)m
 |
| Head to head telomeric sequences (blue) at a fusion point (several possibilities), flanked by proximal (orange, green) and distal (purple) subtelomeric sequences. |
Such kind of sequence is known in the human genome at the 2q13-q14 locus (Ijdo et al.).
>gi|180516|gb|M73018.1|HUMCHR2A Human ancestral telomeric fusion DNA sequence GTGCCCCGGCGCCACGAGGGCGCTGGCGACCACTGTAAGCAAGAGAGCCCTGCGCCTCTCTGCGCCTGCG CGGCCGGGCCGGCCGGCCGCCTTTGCGATGGCGGAGTTGCGTTCTCCTCAGCACAGACCCGGAGAGCACC GCAGGGCGACCTGCGTTGTCTCTGCACAGATTTCGGTGGTACTGCGAAGGCGGACGAGAGTTCTCCTCAG GTCAGACCCGGGCCGGGCCGGCTAGGATACCGCGAGGCGAGCTGCGTTCTGCTCAGCACAGACCTGGGGG TCACCGTAAAGATGGAGCAGCATTCCCCTAAGCACAGAGGTTGGGGCCACTGCCTGGCTTTGTGACAACT CGGGGCGCATCAACGGTGAATAAAATCTTTCCCGGTTGCAGCCGTGAATAATCAAGGTCAGAGACCAGTT AGAGCGGTTCAGTGCGGAAAACGGGAAAGAAAAAGCCCCTCTGAATCCTGGGCAGCGAGATTATCCCAAA GCAAGGCGAGGGGCTGCATTGCAGGGTGAGGGTGAGGGTGAGGGTGAGGGTTAGGGTTTGGGTTGGGGTT GGGGTTGGGGTTGGGGTAGGGTTGGGGTTTGGGTTGGGGTTAGGGTTAGGGGTAGGGGTAGGGTCAGGGT CAGGGTCAGGGTTAGGGTTTTAGGGTTAGGGTTAGGGTTAAGGTTTGGGGTTGGGGTTGGGGTTGGGGTT AGGGGTTAGGGGTTAGGGGTTAGGGTTGGGGTTGGGGGTTGGGGTTGGGGTTAGGGGTAGGGGTAGGGGT AGGGTTAGGGTTAGGGTTAGGGTAAGGGTTAAGGGTTGGGGTTGGGGTTGGGGTTAGGGTTAGGGGTTAG GGTTAGCTAACCCTAACCCTAACCCCTAACCCCTAACCCCAACCCAAACCCCAACCCCAACCCCAACCCT ACCCCTACCCCTAACCCCAACCCTTAACCCTTAACCCTTAACCCTTACCCTAACCCTAACCCAAACCCTA ACCCTAACCCTACCCTAACCCAACCCTAACCCTAACCCTACCCTAAGCCTAAAACCCTAAAACCGTGACC CTGACCTTGACCCTGACCCTTAACCCTTAACCCTTAACCCTAACCCTAACCATAACCCTAAACCCTAACC CTAAACCCTAACCCTACCCTAACCCCAACCCCTAACCCTAACCCCTATACCCTAACCCTAACCCTACCCC TACCCCTAACCCCAACCCCAGCCCCAACCCCAACCCTTACCCTAACCCTACCTAACCCTTAACCCTAACC CCTAACCCTAACCCCTAACCCTACCCCAACCCCAAACCCAACCCTAACCCAACCCTAACCCAACCCTAAC CCCTACCCTAACCCCTAACCCTAACCCCTACCCTAACCCCTAACCCTAACCCCTACCCTAACCCCTAACC CTAGCCCTAGCCCTAACCCTAACCCTCACCCTAACCCTCACCCTAACCCTCACCCTCACCCTCACCCTCA CCCTAACCCAACGTCTGTGCTGAGAAGAATGCTCGTCCGCCTTTAAGGTGCCCCCCAGGTCTGTGCTGAA CAGAACGCACGTCCGCCGTCGCAGTGCCCTCAGCCCGGGTCTGACCTGAGAAGAACTCTGCTCCGCCTTC GCAATAGCCCCGAAGTCTGTGCAGAGGAGAACGCAGCTCCGCCCTCGCGATGCTCTTCGGCTGTGTGCTA AAGAGAACGCAACTCCGCCCTCGCAAAGGCGGCGCCGCCGCGGAGGCCGGAGAGGCGCGGCGCCGCGGAG GCCGGAGAGGCGCGGCGCGCGGGAGGCCGGAGAGGCGCGGCGCCGCGGAGGCCGGAGAGGCGCGGCGCCG CGGAGGCCGGAGAGGCGCGGCGCCGCGGAGGCCGGAGAGGCGCGGCGCCGCGG
The fusion point of this sequence is:
It is the cicatrix of an ancestral chromosomic fusion occured during karyotype evolution. As the human 2q13-q14 contains a short amount of degenerated telomeric motifs these motifs are degenerated so they cannot be detected by QFISH with (CCCTAA)3 PNA probe (most of the time):
Using Polymerase Chain Reaction and only one (TTAGGG)n primer, would it be is possible to detect head-to-head genomic telomeric sequences possibly resulting from chromosomal fusion ?
..TTAGGGTTAGCTAACCCTAA..
Using Polymerase Chain Reaction and only one (TTAGGG)n primer, would it be is possible to detect head-to-head genomic telomeric sequences possibly resulting from chromosomal fusion ?

| Detection of a genomic head-to-head telomeric sequence by PCR amplification with one (TTAGGG)n primer (Dark Blue big arrow) |
Here was investigated the possibility to detect somatic telomeric fusion occurring in senescent or SV40-transformed fibroblasts by one primer PCR amplification of head-to-head telomeric sequences.(J-P Pommier, unpublished data)
It was found that PCR products ( next called G PCR products) can be produced from genomic human DNA with single TTAGGG primers. It is known that human genome DNA contains head-to-head telomeric motifs and possibly some were generated by chromosomal instability in cultured cells.
Material and method:
The cycle conditions of the first PCR assay were inspired by the paper of Ijdo et al. . As no PCR product was detected with these conditions, the stringency conditions were relaxed by decreasing the annealing temperature, without my lab-book, I cannot bring more precision on the cycling conditions.
Two kinds of assay were performed: G-PCR with (TTAGGG)n primers (n=3 or 5) or C-PCR with (CCCTAA)n primers (n=3 or 5) , both with 100ng of genomic DNA (high molecular weight placenta DNA, primary fibroblasts, SV40 transformed fibroblast pre-crisis/post-crisis, wild type S. cerevisiae genomic DNA was also tested). In a second assay, both G an C primers were used at high (100 nM?) or low (20 nM?) concentration. G3-primer and C3-primer had 5' non telomeric sequence allowing a second PCR round with a non telomeric primer.
In an assay, prior PCR, 1 microgramme of genomic DNA was digested by Hpa II (methylation sensitive) or Msp I restriction endonuclease (NEB) targeting GGCC or by an other endonuclease targeting a TTAA restriction site. A 100ng aliquot was then submitted to G-PCR.
PCR products were analysed in a 2-3% agarose gel electrophoresis, stained with ethidium bromide, with a molecular weight standard in the range 200 to 2000 bp approximatly (possibly molecular weight marker IV, Roche?).
Results:
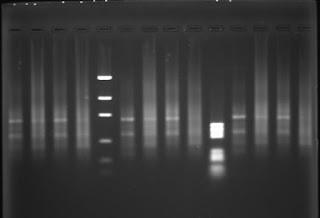
Genomic DNA from placenta, primary fibroblasts, precrisis SV40 transformed fibroblasts submitted to G5-PCR (wells n°6,8,10) yield a smear like PCR product with possibly discrete bands. The same genomic DNA submitted to C5-PCR yield a smears as PCR product with no bands.
C5-PCR or C3-PCR yield a smear as PCR product at high concentration. The PCR product becomes undetectable at low primer concentration. I performed C5-PCR or C3-PCR as a control, I didn't expect to detect a PCR product with a (CCCTAA)n primer due to the head to head orientation of the telomeric motifs in a telomeric fusion. Queue-to-queue telomeric motifs may occur, but those PCR products are likely PCR artifacts.
More interestingly, G5-PCR and G3-PCR yield bands-like PCR products with human DNA in all conditions:
- Bands are observed with human DNA as PCR target, whereas a smear is observed with yeast DNA, indicating that the PCR product detect a specific structure in the human genome which is not present in the yeast genome.
- With the "eyes of faith" (les yeux de la foi), G5-PCR at hight concentration primer, seems to distinguish postcrisis DNA (well 8, upper) from normal or precrisis DNA (wells 6,7 upper) by an relative increase of the amount of the short band. As the SV40 transformed fibroblasts reach the crisis, the frequency of dicentric chromosomes increase and become stable when an immortal clone emerge from the crisis. It is consistent with an accumulation of head-to-head telomeric motif generated by chromosomal fusion between short telomeres.
- The shortest band (~600 bp) vanishes from the PCR product as the G5 primer concentration is diminished regardless the source of DNA (primary cells, precrisis or postcrisis cells). There are possibly several ancestral HTH telomeric motifs per cell, whereas there is less than one neo HTH telomeric motif per cell (statistically around the crisis, cells have 0 or 1 dicentric chromosomes, even some cells have more). As neo HTH telomeric motifs are rare events, decreasing G5 primer would decrease PCR detection efficiency. The same for G3 primer due to thermal stability.
In the last assay the presence of palindromic sequences in potential HTH telomeric motifs was tested with G5-PCR only.
Some of them generate palindromic sequences as GGCC cut by the isoschisomeres Hpa II / Msp I restriction endonucleases, or TTAA .There are 36 possible head-to-head telomeric motifs as follow:
 |
| Head-to-head telomeric motif generation by chromosomic fusion |
Some of them generate palindromic sequences as GGCC cut by the isoschisomeres Hpa II / Msp I restriction endonucleases, or TTAA .There are 36 possible head-to-head telomeric motifs as follow:
TTAGGG
|
TAGGGT
|
AGGGTT
|
GGGTTA
|
GGTTAG
|
GTTAGG
|
|
CCCTAA
|
TTAGGGCCCTAA
|
TAGGGTCCCTAA
|
AGGGTTCCCTAA
|
GGGTTACCCTAA
|
GGTTAGCCCTAA
|
GTTAGGCCCTAA
|
CCTAAC
|
TTAGGGCCTAAC
|
TAGGGTCCTAAC
|
AGGGTTCCTAAC
|
GGGTTACCTAAC
|
GGTTAGCCTAAC
|
GTTAGGCCTAAC
|
CTAACC
|
TTAGGGCTAACC
|
TAGGGTCTAACC
|
AGGGTTCTAACC
|
GGGTTACTAACC
|
GGTTAGCTAACC
|
GTTAGGCTAACC
|
TAACCC
|
TTAGGGTAACCC
|
TAGGGTTAACCC
|
AGGGTTTAACCC
|
GGGTTATAACCC
|
GGTTAGTAACCC
|
GTTAGGTAACCC
|
AACCCT
|
TTAGGGAACCCT
|
TAGGGTAACCCT
|
AGGGTTAACCCT
|
GGGTTAAACCCT
|
GGTTAGAACCCT
|
GTTAGGAACCCT
|
ACCCTA
|
TTAGGGACCCTA
|
TAGGGTACCCTA
|
AGGGTTACCCTA
|
GGGTTAACCCTA
|
GGTTAGACCCTA
|
GTTAGGACCCTA
|
If the telomeric fusion point contains a palindromic sequence, submitting the genomic DNA to endonuclease prior G5-PCR may prevent the PCR amplification:
 |
| Endonuclease digestion of DNA may prevent G-PCR amplification. |
In uncut DNA (primary fibroblasts (line 1), precrisis fibroblasts (line 6), immortalized clone (line 11)), G5-PCR produces two major bands bellow 1200 bp. Those bands vanished as DNA is cut with MspI (GGCC) or with an endonuclease targeting TTAA sequence (from left to right, primary cells, pre or post-crisis fibroblasts:lines 2, 4, 7,9, 12, 13), indicating that palindromic sequences exist between inverse tandem telomeric motifs.
When genomic DNA is cut with HpaII, an endonuclease cutting GGCC motifs free of methyl-cytosine, the G-PCR product cannot be distinguished from the uncut DNA (lines 3, 8, 13), indicating that the cytosines are methylated in/between inverse telomeric motifs.
If the aim was to detect somatic telomeric fusions associated to cellular senescence or to cellular transformation, the G5-PCR assays failed to distinguish clearly the different cell states. The G-PCR to detect end-to-end telomeric fusion may be optimized (PCR optimization) by increasing stringency:
- by increasing annealing T°
- by decreasing elongation time
- using hot-start PCR (Taq hot-start or wax)
Polyacrylamide gel electrophoresis (PAGE) with P32-labelled (TTAGGG)n primer instead of ethidium-bromide stained agarose gel to visualize PCR product, may detect somatic telomeric fusions in the range 0-600 bp. In the last example, the six last lines correspond to G-PCR products were visualized by PAGE (may be products corresponding to lines 6 to 11 in the second assay).
Real time PCR, in-situ PCR on nuclei could be also considered.
 |
| Six G5-PCR products (right side) in a PAGE+Ethidium Bromide staining |
This last hypothesis could be checked by FISH. The G-PCR product could be labelled (after a second round of PCR) with Fluo-dNTP and hybridized on metaphasic chromosomes. Three FISH conditions could be set:
- labelled G-PCR product alone: some locci should be detected.
- labelled G-PCR product+excess of Cot1 DNA: should compete satellite DNA. Detected locci should contain non satellite sequence, possibly telomeres or unique sequences.
- labelled G-PCR product+excess of unlabeled telomeric DNA (produced by PCR, Ijdo & al.): detected locci should not contain telomeric motifs.


 ,
,