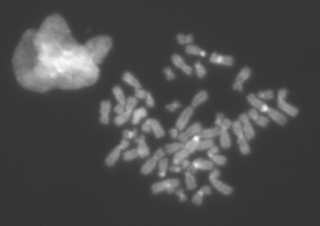
 |
| Part of the original 16bits TIF image |
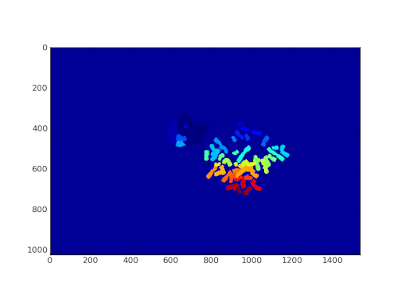 |
| Labelled image displayed with pylab |
The mean gray level value is also calculated for each labelled region with pymorph.grain:
[ 1301.88575849 23.00804348 704.34775673 629.5625966 1410.2556391
871.05686546 946.11368209 841.65226782 860.98429319 1526.48712121
705.22321429 608.3359841 929.86325301 858.11372549 1000.02900378
831.46130952 717.89433962 790.23236741 919.48754448 603.34122288
830.35550936 1325.0239521 843.45842697 880.9575 864.25319693
970.12876254 901.85921959 914.75644699 902.17860963 864.04659498
960.92638037 892.95342466 946.98757358 1119.16554163 835.44263629
872.87977369 838.04045734 734.5129683 1147.49576784 995.61397059
839.4835443 823.52125984 795.88679245 898.91109422 748.07751938
635.60344828]
the python code used for that result:
from scipy import ndimage as ND
import scipy
import numpy as np
import readmagick
import mahotas
import pymorph
import pylab
import os
def SegmentMetaphase(image):
#print "def image:",image.mean()
#print "def background:",back.mean()
#print "def noback:",im.mean()
noBack=RemoveModalBackground(image)
print "from Segmeta noBack:",noBack.min(),noBack.mean()
blurLowRes=ND.filters.gaussian_filter(noBack,13)
blurHiRes=ND.filters.gaussian_filter(noBack,1)
midPass=pymorph.subm(blurHiRes,0.70*blurLowRes)
bin=(midPass>1.5*midPass.mean())
binLowRes=pymorph.open(bin,pymorph.sedisk(4))
binLR=pymorph.edgeoff(binLowRes)
blur=ND.filters.gaussian_filter(noBack,9)
hiPass=pymorph.subm(noBack,1.0*blur)
gradLowRes=pymorph.gradm(noBack)
#build seeds for watershed
bSeeds1=(hiPass>hiPass.mean())
bSeeds2=scipy.logical_and(bSeeds1,binLR)
#bSeeds3=ND.binary_fill_holes(bSeeds2)
bSeeds4=ND.binary_opening(bSeeds2,iterations=4)
outline=mahotas.bwperim(bin, n=4)
#display=pymorph.overlay(image,outline)#Hugly
markers,nr_obj=ND.label(bSeeds4)
labeled=mahotas.cwatershed(gradLowRes, markers, Bc=None, return_lines=False)
return labeled
def RemoveModalBackground(image):
mode=ModalValue(image)
back = np.zeros(image.shape, image.dtype)
back.fill(mode)
#print "def background:",back.mean()
im=pymorph.subm(image,back)
return im
def ModalValue(image):
'''look for the modal value of an image'''
#print image.dtype
if image.dtype=="uint8":
depthmax=255
print "8bits"
if image.dtype=="uint16":
depthmax=65535
print "16bits"
histo=mahotas.fullhistogram(image)
countmax=histo.max()
print "countmax:",countmax
print "image max",image.max()
mig=image.min()#image min graylevel
mag=image.max()#image max gray level
mode=0
countmax=0#occurence of a given grayscale
print "mig=",mig," mag=",mag
for i in range(mig,mag-1,1):
test=histo[i]>countmax
#print "test:",test,"histo(",i,")=", histo[i],"max",countmax
if test:
countmax=histo[i]
mode=i
#print "mode",mode
return mode
def BorderKill(imlabel):
'''remove labelled objects touching the image border'''
#from pythonvision.org
#pymorph.edgeoff should do the same
whole = mahotas.segmentation.gvoronoi(imlabel)
borders = np.zeros(imlabel.shape, np.bool)
borders[ 0,:] = 1
borders[-1,:] = 1
borders[:, 0] = 1
borders[:,-1] = 1
at_border = np.unique(imlabel[borders])
for obj in at_border:
whole[whole == obj] = 0
return whole
user=os.path.expanduser("~")
workdir=os.path.join(user,"Applications","ImagesTest","JPP60-3","DAPI")
file="1.tif"
complete_path=os.path.join(workdir,file)
if __name__ == "__main__":
#print complete_path
dapi=readmagick.readimg(complete_path)
readmagick.writeimg(dapi,os.path.join(workdir,"dapi.png"))
NoBackIm=RemoveModalBackground(dapi)
dapilab1=SegmentMetaphase(dapi)
readmagick.writeimg(dapilab1,os.path.join(workdir,"dapilabel.png"))
print pymorph.grain(NoBackIm,dapilab1, 'mean', option='data')
pylab.imshow(dapilab1)
pylab.show()





